This tutorial corresponds to 03_custom_dataset_mlp folder in the source code.
We have prepared your own dataset, MyDataset, in previous post. Training procedure for this dataset is now almost same with MNIST traning.
Differences from MNIST dataset are,
- This task is regression task (estimate final “value”), instead of classification task (estimate the probability of category)
- Training data and validation/test data is not splitted in our custom dataset
Model definition for Regression task training
Our task is to estimate the real value “t” given real value “x“, which is categorized as regression task.
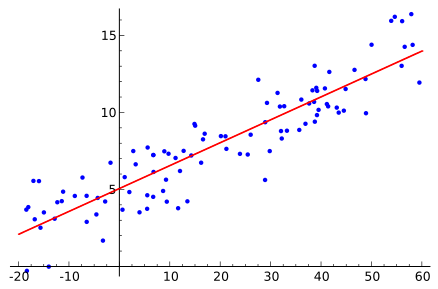
We often use mean squared error as loss function, namely,
$$ L = \frac{1}{D}\sum_i^D (t_i – y_i)^2 $$
where \(i\) denotes i-th data, \(D\) is number of data, and \(y_i\) is model’s output from input \(x_i \).
The implementation for MLP can be written as my_mlp.py,
class MyMLP(chainer.Chain):
def __init__(self, n_units):
super(MyMLP, self).__init__()
with self.init_scope():
# the size of the inputs to each layer will be inferred
self.l1 = L.Linear(n_units) # n_in -> n_units
self.l2 = L.Linear(n_units) # n_units -> n_units
self.l3 = L.Linear(n_units) # n_units -> n_units
self.l4 = L.Linear(1) # n_units -> n_out
def __call__(self, *args):
# Calculate loss
h = self.forward(*args)
t = args[1]
self.loss = F.mean_squared_error(h, t)
reporter.report({'loss': self.loss}, self)
return self.loss
def forward(self, *args):
# Common code for both loss (__call__) and predict
x = args[0]
h = F.sigmoid(self.l1(x))
h = F.sigmoid(self.l2(h))
h = F.sigmoid(self.l3(h))
h = self.l4(h)
return h
In this case, MyMLP model will calculate y (target to predict) in forward computation, and loss is calculated at __call__ function of the model.
Data separation for validation/test
When you are downloading publicly available machine learning dataset, it is often separated as training data and test data (and sometimes validation data) from the beginning.
However, our custom dataset is not separated yet. We can split the existing dataset easily with chainer’s function, which includes following function
chainer.datasets.split_dataset(dataset, split_at, order=None)chainer.datasets.split_dataset_random(dataset, first_size, seed=None)chainer.datasets.get_cross_validation_datasets(dataset, n_fold, order=None)chainer.datasets.get_cross_validation_datasets_random(dataset, n_fold, seed=None)
refer SubDataset for details.
These are useful to separate training data and test data, example usage is as following,
# Load the dataset and separate to train data and test data
dataset = MyDataset('data/my_data.csv')
train_ratio = 0.7
train_size = int(len(dataset) * train_ratio)
train, test = chainer.datasets.split_dataset_random(dataset, train_size, seed=13)
Here, we load our data as dataset (which is subclass of DatasetMixin), and split this dataset into train and test using chainer.datasets.split_dataset_random function. I split train data 70% : test data 30%, randomly in above code.
We can also specify seed argument to fix the random permutation order, which is useful for reproducing experiment or predicting code with same train/test dataset.
Training code
The total code looks like, train_custom_dataset.py
from __future__ import print_function
import argparse
import chainer
import chainer.functions as F
import chainer.links as L
from chainer import training
from chainer.training import extensions
from chainer import serializers
from my_mlp import MyMLP
from my_dataset import MyDataset
def main():
parser = argparse.ArgumentParser(description='Train custom dataset')
parser.add_argument('--batchsize', '-b', type=int, default=10,
help='Number of images in each mini-batch')
parser.add_argument('--epoch', '-e', type=int, default=20,
help='Number of sweeps over the dataset to train')
parser.add_argument('--gpu', '-g', type=int, default=-1,
help='GPU ID (negative value indicates CPU)')
parser.add_argument('--out', '-o', default='result',
help='Directory to output the result')
parser.add_argument('--resume', '-r', default='',
help='Resume the training from snapshot')
parser.add_argument('--unit', '-u', type=int, default=50,
help='Number of units')
args = parser.parse_args()
print('GPU: {}'.format(args.gpu))
print('# unit: {}'.format(args.unit))
print('# Minibatch-size: {}'.format(args.batchsize))
print('# epoch: {}'.format(args.epoch))
print('')
# Set up a neural network to train
# Classifier reports softmax cross entropy loss and accuracy at every
# iteration, which will be used by the PrintReport extension below.
model = MyMLP(args.unit)
if args.gpu >= 0:
chainer.cuda.get_device(args.gpu).use() # Make a specified GPU current
model.to_gpu() # Copy the model to the GPU
# Setup an optimizer
optimizer = chainer.optimizers.MomentumSGD()
optimizer.setup(model)
# Load the dataset and separate to train data and test data
dataset = MyDataset('data/my_data.csv')
train_ratio = 0.7
train_size = int(len(dataset) * train_ratio)
train, test = chainer.datasets.split_dataset_random(dataset, train_size, seed=13)
train_iter = chainer.iterators.SerialIterator(train, args.batchsize)
test_iter = chainer.iterators.SerialIterator(test, args.batchsize, repeat=False, shuffle=False)
# Set up a trainer
updater = training.StandardUpdater(train_iter, optimizer, device=args.gpu)
trainer = training.Trainer(updater, (args.epoch, 'epoch'), out=args.out)
# Evaluate the model with the test dataset for each epoch
trainer.extend(extensions.Evaluator(test_iter, model, device=args.gpu))
# Dump a computational graph from 'loss' variable at the first iteration
# The "main" refers to the target link of the "main" optimizer.
trainer.extend(extensions.dump_graph('main/loss'))
# Take a snapshot at each epoch
#trainer.extend(extensions.snapshot(), trigger=(args.epoch, 'epoch'))
trainer.extend(extensions.snapshot(), trigger=(1, 'epoch'))
# Write a log of evaluation statistics for each epoch
trainer.extend(extensions.LogReport())
# Print selected entries of the log to stdout
# Here "main" refers to the target link of the "main" optimizer again, and
# "validation" refers to the default name of the Evaluator extension.
# Entries other than 'epoch' are reported by the Classifier link, called by
# either the updater or the evaluator.
trainer.extend(extensions.PrintReport(
['epoch', 'main/loss', 'validation/main/loss', 'elapsed_time']))
# Plot graph for loss for each epoch
if extensions.PlotReport.available():
trainer.extend(extensions.PlotReport(
['main/loss', 'validation/main/loss'],
x_key='epoch', file_name='loss.png'))
else:
print('Warning: PlotReport is not available in your environment')
# Print a progress bar to stdout
trainer.extend(extensions.ProgressBar())
if args.resume:
# Resume from a snapshot
serializers.load_npz(args.resume, trainer)
# Run the training
trainer.run()
serializers.save_npz('{}/mymlp.model'.format(args.out), model)
if __name__ == '__main__':
main()
[hands on]
Execute train_custom_dataset.py to train the model. Trained model parameter will be saved to result/mymlp.model.